Abstract
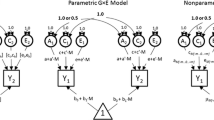
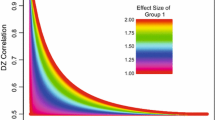
It is likely that all complex behaviors and diseases result from interactions between genetic vulnerabilities and environmental factors. Accurately identifying such gene–environment interactions is of critical importance for genetic research on health and behavior. In a previous article we proposed a set of models for testing alternative relationships between a phenotype (P) and a putative moderator (M) in twin studies. These include the traditional bivariate Cholesky model, an extension of that model that allows for interactions between M and the underling influences on P, and a model in which M has a non-linear main effect on P. Here we use simulations to evaluate the type I error rates, power, and performance of the Bayesian Information Criterion under a variety of data generating mechanisms and samples sizes (n = 2,000 and n = 500 twin pairs). In testing the extension of the Cholesky model, false positive rates consistently fell short of the nominal Type I error rates (\( \alpha = 10,.05,.01 \)). With adequate sample size (n = 2,000 pairs), the correct model had the lowest BIC value in nearly all simulated datasets. With lower sample sizes, models specifying non-linear main effects were more difficult to distinguish from models containing interaction effects. In addition, we provide an illustration of our approach by examining possible interactions between birthweight and the genetic and environmental influences on child and adolescent anxiety using previously collected data. We found a significant interaction between birthweight and the genetic and environmental influences on anxiety. However, the interaction was accounted for by non-linear main effects of birthweight on anxiety, verifying that interaction effects need to be tested against alternative models.


Similar content being viewed by others
Notes
Stata and Mplus scripts for data generation and model fitting are available from the first author at.http://www.waisman.wisc.edu/twinresearch/researchers/vanhullecv.shtml
References
Asbury K, Dunn JF, Plomin R (2006) Birthweight-discordance and differences in early parenting relate to monozygotic twin differences in behaviour problems and academic achievement at age 7. Dev Sci 9(2):F22–F31. doi:10.1111/j.1467-7687.2006.00469.x
Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J et al (2011) OpenMx: an open source extended structural equation modeling framework. Psychometrika 76(2):306–317. doi:10.1007/s11336-010-9200-6
Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M (2001) Exploring gene–environment interactions: socioregional moderation of alcohol use. J Abnorm Psychol 110(4):625–632. doi:10.1037/0021-843X.110.4.625
Johnson W, Krueger RF (2005) Higher perceived life control decreases genetic variance in physical health: evidence from a national twin study. J Pers Soc Psychol 88(1):165–173. doi:10.1037/0022-3514.88.1.165
Johnson W, McGue M, Iacono WG (2009) School performance and genetic and environmental variance in antisocial behavior at the transition from adolescence to adulthood. Dev Psychol 45:973–987
Lau JY, Eley TC (2008) Disentangling gene-environment correlations and interactions on adolescent depressive symptoms. J Child Psychol Psy 149:142–150
Lahey BB, Waldman ID, Loft JD, Hankin B, Rick J (2004) The structure of child and adolescent psychopathology: generating new hypotheses. J Abnorm Psychol 113:358–385
McArdle JJ, Goldsmith HH (1990) Alternative common factor models for multivariate biometric analyses. Behav Genet 20(5):569–608. doi:10.1007/BF01065873
Medland SE, Neale MC, Eaves LJ, Neale BM (2008) A note on the parameterization of Purcell’s G × E model for ordinal and binary data. Behav Genet 39(2):220–229. doi:10.1007/s10519-008-9247-7
Muthén L, Muthén B (1998) Mplus user’s guide, vol 6. Muthén & Muthén, Los Angeles
Neale M, Cardon L (1992) Methodology for genetic studies of twin and families. NATO ASI Series D: behavioral and social sciences, vol 67. Kluwer Academic Publishers, Boston
Price T, Jaffee S (2008) Effects of the family environment: gene–environmentinteraction and passive gene–environmentcorrelation. Dev Psychol 44(2):305–315. doi:10.1037/0012-1649.44.2.305
Purcell S (2002) Variance components models for gene–environment interaction in twin analysis. Twin Res 5(6):554–571. doi:10.1375/136905202762342026
R Development Core Team. (2011). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from http://www.r-project.org
Raftery AE (1995) Bayesian model selection in social research. Sociol Methodol 25:111–163
Rathouz PJ, Van Hulle CA, Rodgers JL, Waldman ID, Lahey BB (2008) Specification, testing, and interpretation of gene-by-measured-environment interaction models in the presence of gene–environment correlation. Behav Genet 38(3):301–315. doi:10.1007/s10519-008-9193-4
Saunders C, Bishop D, Barrett J (2003) Sample size calculations for main effects and interactions in case–control studies using Stat’s nchi2 and npnchi2 functions. Stata J 3(1):47–56
Silventoinen K, Hasselbalch AL, Lallukka T, Bogl L, Pietilainen KH, Heitmann BL, Schousboe K et al (2009) Modification effects of physical activity and protein intake on heritability of body size and composition. Am J Clin Nutr 90(4):1096–1103. doi:10.3945/ajcn.2009.27689
South SC, Krueger RF (2008) Marital quality moderates genetic and environmental influences on the internalizing spectrum. J Abnorm Psychol 117(4):826–837. doi:10.1037/a0013499
StataCorp. (2011). Stata Statistical Software: Release 12. College Station, TX: StataCorp LP
Turkheimer E, Haley A, Waldron M, D’Onofrio B, Gottesman II (2003) Socioeconomic status modifies heritability of IQ in young children. Psychol Sci 14(6):623–628. doi:10.1046/j.0956-7976.2003.psci_1475.x
van Beijsterveldt T, Boomsma DI (2008) An exploration of gene–environment interaction and asthma in a large sample of 5-year-old Dutch twins. Twin research and human genetics 11(2):143–149
Acknowledgments
This study was funded by the NIH Grant R21 MH086099 from the National Institute for Mental Health. Infrastructure support was provided by the Waisman Center via a core grant from the National Institute of Child Health and Human Development (P30 HD03352).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Stacey Cherny.
Appendices
Appendix 1
Estimated sample size analysis
We followed the method recommended by Saunders et al. (2003). Assume \( \widehat{E}(\chi^{2} ) = Q \)for N simulations, where \( \widehat{E}(\chi^{2} ) \) is the mean LRT between HA and H0 across simulations. Then, let \( \frac{Q - P}{N} = \hat{k} \) where p is the degrees of freedom for the LRT. From Q and \( \hat{k} \), we can compute the χ2 non-centrality parameter for a given sample size \( \bar{N} \) and thereby generate power. Reversing this process, given p and a target power \( (1 - \beta ) \), we can, via the non-centrality parameter \( \lambda_{p,1 - \beta } \), compute the sample size \( \bar{N} \) needed to achieve power \( (1 - \beta ) \). If we set \( \lambda_{p,1 - \beta } = \hat{k}\bar{N} \) then
The non-centrality parameter was calculated using Stata’s npnchi2 function (Stata Corp. 2009 Stata Statistical Sofware: Release 11. College Station, TX: StataCorp, LP) \( {{\uplambda}}_{{{\text{p,1}} - \beta }} {\text{ = npnchi2(p,invchi2(p,(1}} - \alpha ) ) , ( 1- (\beta / 1 0 0 ) / {\text{Q)}} \).
Appendix 2
See (Table 6)
Appendix 3
See (Table 7)
Rights and permissions
About this article
Cite this article
Van Hulle, C.A., Lahey, B.B. & Rathouz, P.J. Operating Characteristics of Alternative Statistical Methods for Detecting Gene-by-Measured Environment Interaction in the Presence of Gene–Environment Correlation in Twin and Sibling Studies. Behav Genet 43, 71–84 (2013). https://doi.org/10.1007/s10519-012-9568-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-012-9568-4